| Jump to section |
About HGFDB | User Manual | Funding | Contract us |
About HGFDB
Helmeted guinea fowl(HGF; Numida meleagris) is vigorous, hardy, and mostly disease-free game birds. The rearing of HGF is a potential alternative poultry system. Because of the characteristics of the HGF, the study of HGF is valuable, so we conduct an improved domesticated HGF genome de novo assembly generated by long and short read sequencing together with optical and chromatin interaction mapping. Using this assembly as the reference, we performed population genomic analyses for newly sequenced whole-genomes for 129 birds including domesticated animals, wild progenitors, and closely related wild species. The data are important to provide valuable genetic resources for researchers and breeders to improve production in this species. For this purpose, we have developed a comprehensive and easy-to-use web-based database, HGFDB. The HGFDB harvests 129 genomes from 89 domestic HGF, 34 wild HGF, five vulturine guinea fowl (Acryllium vulturinum), and one crested guinea fowl (Guttera pucherani). The database also contains 15173 protein-coding genes, in which 14373 were functional annotated and provides search functionality, a BLAST server, JBrowse and genome and resquencing data information etc. All data in HGFDB are freely available for download.
User Manual
Database overview
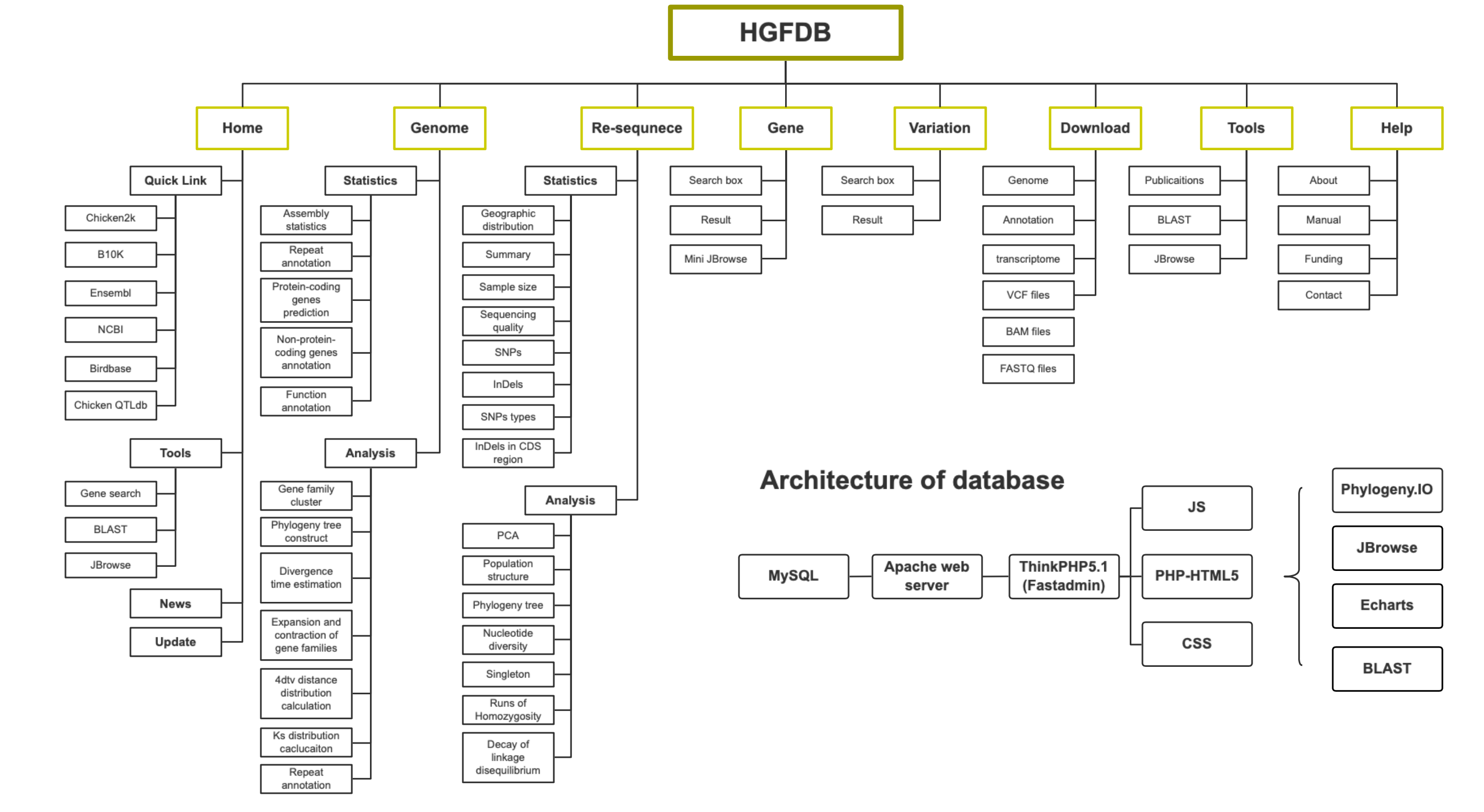
Statistics and Analysis
In the current version of 2020-08, both the Statistics and Analysis pages contain two sub-pages, Genome and Re-sequence, contains multi-angle data analysis and statistics through progressive functions, interactive graphical interface and Multiple data visualization components, most of the figures in this page are dynamic and interactive, and users can understand the current group information more clearly, Change the style or browse the data structure through the toolbar. The phylogeny is implemented by phylogeny.IO.(https://github.com/oist/phylogeny-io). Other dynamic interactive charts are implemented by Echarts(https://echarts.apache.org/zh/index.html).
Next, we use the section of Population structure under the Re-sequence page as an example to help users use functional components more efficiently.(Figure attached below)

1.Mouse click legend: Hovering the mouse over the legends will display all sample information for that color. Clicking on the figure legend will hide components represented by the corresponding color in the plot, and re-clicking on the inactive color (shown in grey) will enable it show again on the plot.
2.Mouse hover event: Hovering on an element in a plot will display an information box containing detailed data about this element.
3.Mouse wheel scroll or drag: This behavior will zoom in or out of the corresponding area in the figure, or place aim content the area in the center of the field of view.
4.Some buttons will allow you to browse the data format, download image and restore the image to default.
5.Filter criteria box: This section will allow you to filter the data in the chart by a certain classification or change the data set.
Search
We set up search tools in Gene and Variation mode that can search according to the conditions, Variation type choice you need query variable types (SNP or Indel), Chr choose chromosome number, you need to search Pos set query target location on chromosome interval (start to end). you can search also through the gene name, the gene name search box type in the name of the gene you need query and five public databases were used for annotation, including GO, KEGG, InterPro, Swissprot and TrEMBL, enter gene name such as AGL, TYP, SLC6A4, FMO5, PLXNA4 etc., In the search results, you will see the target's chromosome number, specific sites, specific mutant bases and related information. In the current version of 2020-01, Variation contains 44,035,924 non-redundant SNPs and 4,214,076 InDels of 129 genomes.
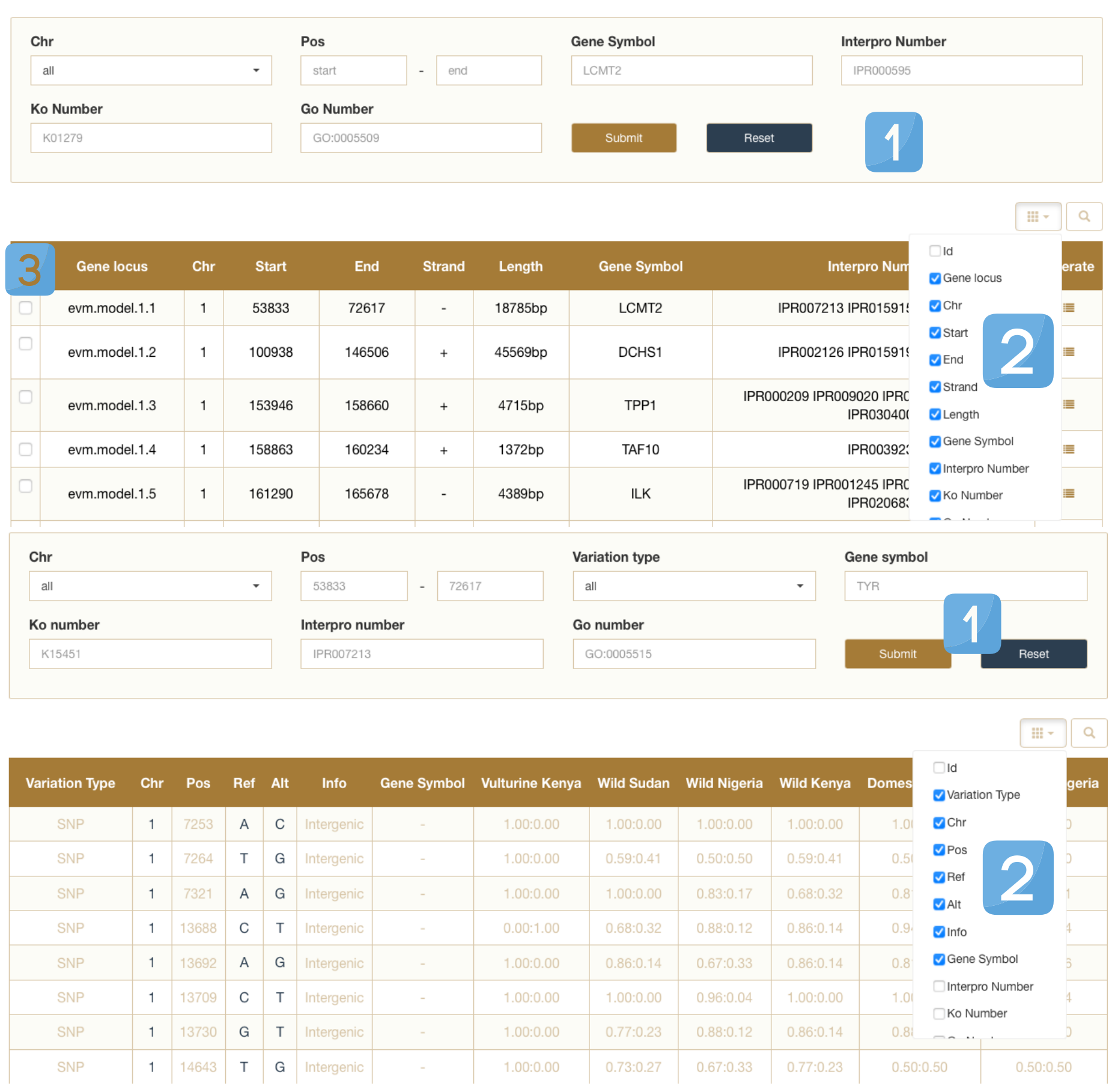
1.Multi-condition search box.
2.Filter the components that display information.
3.In the Gene module, you can check the item to locate in the jbrowse below.
BLAST
NCBI's BLAST is also available in our database as an online tool, which can be found under the "Tools" menu. User can enter query sequences of DNA against DHGF genome database getting a group of high-scoring pairwise alignments.

JBrowse
JBrowse (Skinner,M.E et al.,2009) is the next genome browser, which supports easy navigation and allows the uploading of local files in many formats to complement the web system data. JBrowse is JavaScript-based software; it loads multiple tracks quickly and scales well for large regions.
HGFDB provides JBrowse-based integration of guinea fowl genome annotation, including gene, CDS, mRNA. User can easily browse interested regions in the genome.

1.Filter the tracks displayed.
2.Target positioning through tool components.
3.Swipe to view target information or click to view detailed information.
Download
We designed a dedicated page so that user can quickly download all the data they need. This page include assembled reference genome sequences of the HGF, genome annotation, SNP variation, transcriptome sample information and transcriptome to visualize locally. In addition, if user want any data that we aren’t update, please contrast us.
Funding
This work was supported by Digitalization of Biological Resource Project (No. 202002AA100007), Yunnan.
Contract us
Laboratory: State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan
Institute: Yunnan Agricultural University
Address: No. 21 Qingsong Road, Ciba, Kunming 650204, Yunnan Province, China
E-mail: HGFDB@dongyang-lab.org